Description/Help)
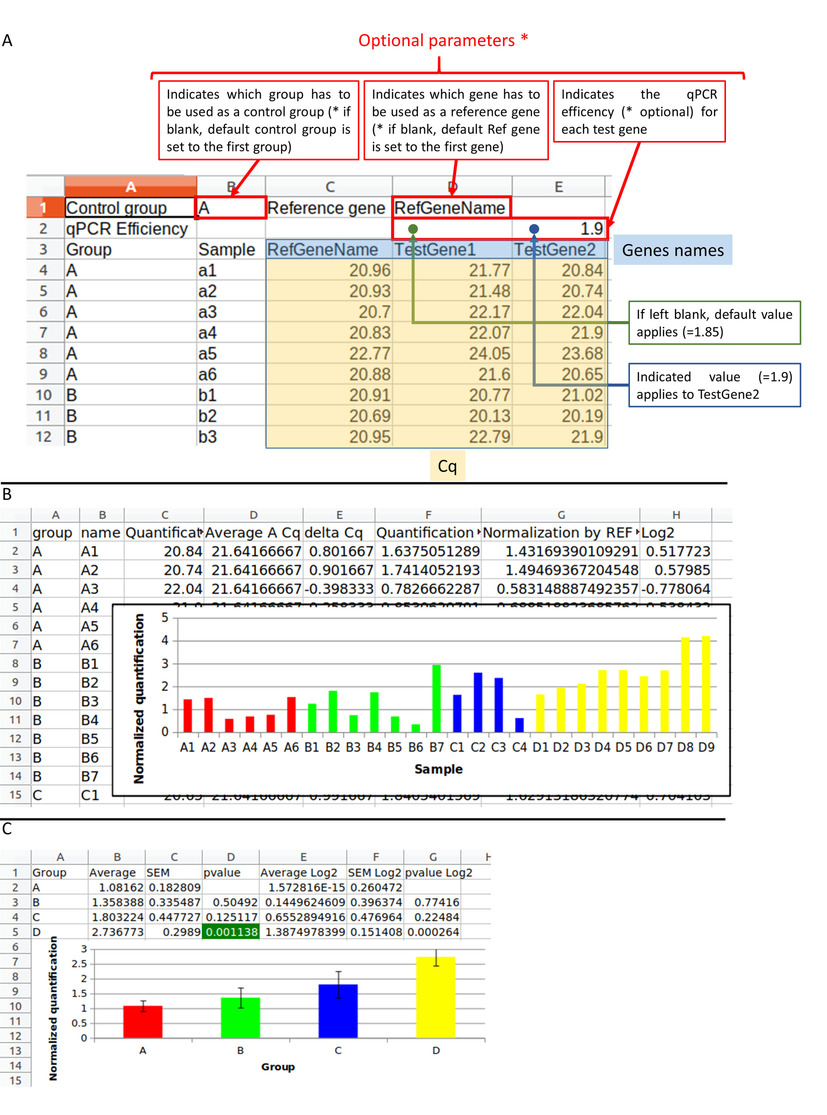
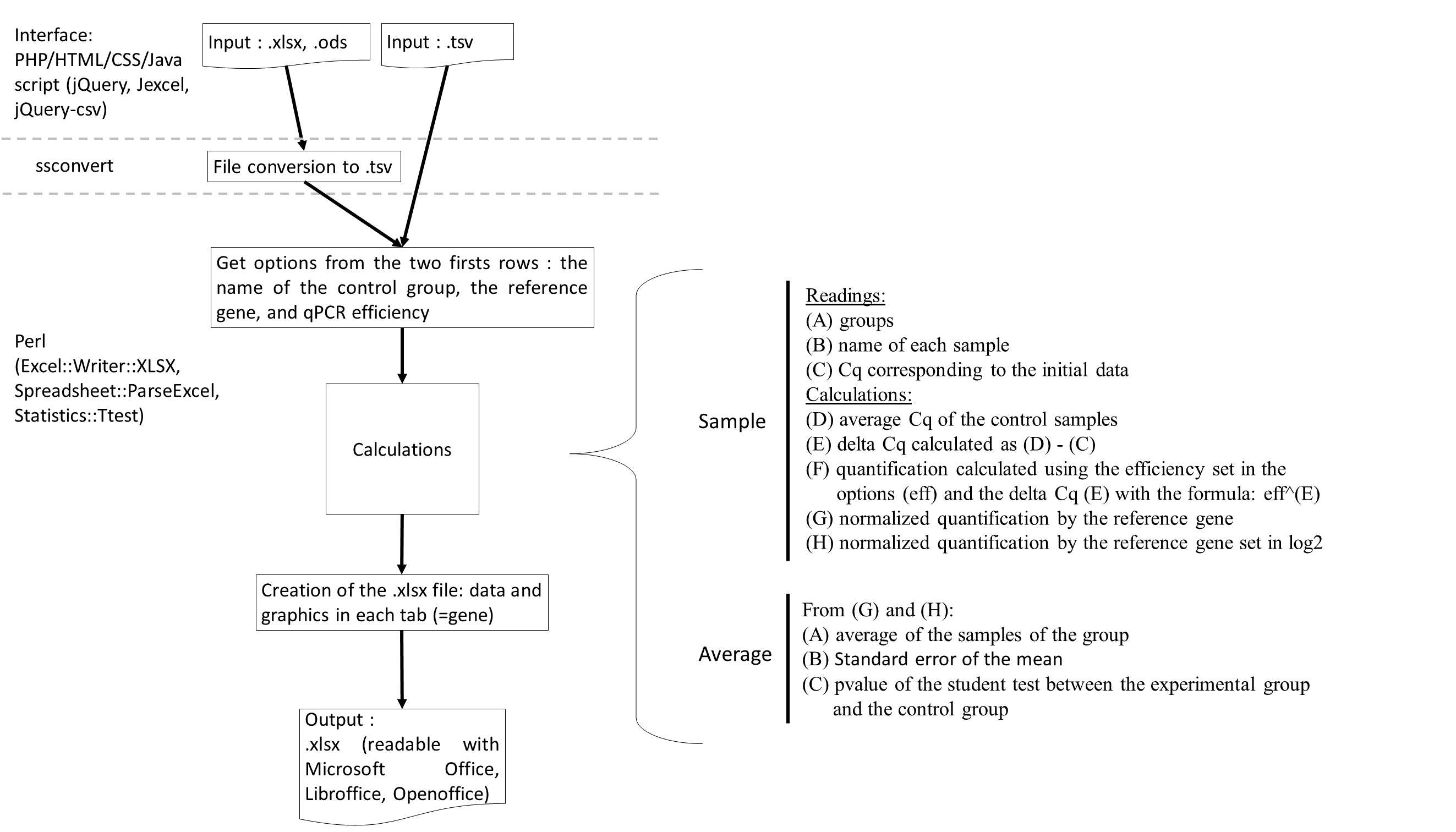
A) The input file has to be in .tsv, .xls, .xlsx. or .odt format. The first two lines are optional. The first defines the control group in B1 cell : the "A" group is choiced. The reference gene in D1 cell : the "RefGeneName" gene is selected. The second allows to define the qPCR efficiency for each gene written on the 3rd line. On the 3rd line there are 2 column headers : "Group" and "Sample" and then there are the genes: "RefGeneName", "TestGene1", "TestGene2". The other rows correspond to the data table : sample according to the Cq. Submitting this file to "do my qPCR calculation" allows you to obtain the result file in Excel format. B) The Excel file contains the normalized Cq for each sample with histograms. C) The Excel file also contains the average results and the student test for each experimental group between the control group.

 1Université Clermont Auvergne, INRA, UNH, Unité de Nutrition Humaine, CRNH Auvergne, Clermont-Ferrand, France
2Université Clermont Auvergne, VetAgro Sup, INRA, UMR1213, Unité Mixte de Recherche sur les Herbivores, Clermont-Ferrand, France
1Université Clermont Auvergne, INRA, UNH, Unité de Nutrition Humaine, CRNH Auvergne, Clermont-Ferrand, France
2Université Clermont Auvergne, VetAgro Sup, INRA, UMR1213, Unité Mixte de Recherche sur les Herbivores, Clermont-Ferrand, France